Wednesday, April 19, 2017
TCUP -- HRAM-MS + Free code = bacterial identifications!!
I'll be clear -- this isn't the logo for the new free TCUP methodology/software that I'm very excited about this morning! This is an unrelated product that I'm very excited about.
This is TCUP and this paper hasn't been edited for print yet, but it is still very much worth looking at. You can find the early release here! (Sorry, traveling and bent my laptop a little. Can't figure out how to right click and insert link...) EDIT: Got it!!
We've seen stuff like this before over the years, right? I mean, the great Catherine Fenselau wrote a paper called "identifying bacteria with mass spectrometry" in 1975! What makes this one special?
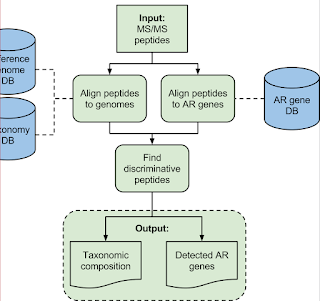
1) High res accurate mass fragment ions (QE Classic!)
2) A really smart discriminatory (open...but Python...) data processing pipeline that can take genomic information that we know for sure (perfectly curated peptide/protein sequences) and even stuff we don't know -- by using BLAT or BLAST (think of BLAT as a better BLAST that only works for people who aren't using Windows)
Here is the goal -- most clinical microbiology labs are still doing stuff the way Louis Pasteur and his students were doing it -- isolate a colony, multiple that colony, see if that colony will grow on this agar plate without glucose or on this agar plate with blood -- and on and on. There are big downsides here:
1) Microbiology labs are super gross. Ever walked into or by a clinical microbiology lab? I had a friend who worked in one when I was in clinical chem. I stopped having lunch with him 'cause if I had to go in there I wouldn't actually want lunch anymore. Gross. But more importantly....
2) Traditional microbiology approaches are SUPER SLOW. You have to wait for a bacteria to grow, isolate, grow isolate, and not everything is Clostridium perfringens (doubles in like 20 minutes without oxygen and insanely stinky!) some of this stuff doesn't make one cell division in A DAY!
Okay -- and I know there is a bunch of MALD-TOF based microbial identification tools out there (that are awesome for the microbiologists! and, p.s., almost completely pioneered at JHU/JHH) but they have limits. They still require pure cultures are isolated. They require matching libraries for the organisms, and so on.
What is awesome about this technique?
1) Mixed cultures? -- no problem (no colony isolations!!) HRAM MS/MS don't care!
2) Deep confident characterization of the organisms in those cultures? Yeah!
3) Can handle the fact that bacteria mutate like CRAZY? Yes! Integration of BLAT/BLAST allows for sequence variation.
We're gonna see lots of this going forward -- but this nice free software package might save the people out there working on this a lot time!
Subscribe to:
Post Comments (Atom)


No comments:
Post a Comment